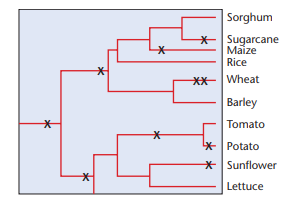
Polyploidy in plants appears to be a significant factor in evolution. However, recent studies (reviewed by Adams and Wendel, 2005) explain that after polyploidization, differential gene silencing and loss reshapes and subfunctionalizes the genome. In the next column are inferred polyploidy events (x) in selected lineages of familiar plants (modified from Blanc and Wolfe, 2004).
(a) Would you expect to see a similar pattern of polyploidization in animal lineages?
(b) In what way would the merging of two genomes (allopolyploidy) support rapid evolution?
(c) Polyploidization in maize occurred approximately 11 million years ago, yet approximately half of all the duplicated genes have been lost. How might polyploidization followed by gene loss contribute to speciation?
(d) Apparently, gene loss is not random. In one study, genes involved in transcription and signal transduction have been retained more often than those involved in DNA repair. In addition, genes for organellar proteins seem to be preferentially lost. How might one explain these findings?
(e) Present a drawing that depicts the union of two different genomes in an initial allopolyploidization event followed by selective gene loss that returns the genome to a genetically diploid amalgam.